Julius Scherer
Development of an Automatic Pathology Recognition System Using Machine Learning on the Basis of Clinical Raw Data from Optical Coherence Tomography
Introduction
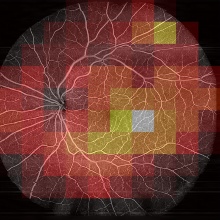
Diabetic Retinopathy (DR) is an eye disease which affects about every third diabetes patient. It is the leading cause of blindness in working-age adults, even though DR complications are preventable if the disease is timely diagnosed. Optical Coherence Tomography (OCT) is an imaging technique, based on light interferometry, that can be used to monitor and diagnose DR. For this, the interferometric data is firstly converted into images and afterwards analysed by medical experts. Machine Learning (ML) techniques have been shown to achieve matching or exceeding DR classification performance compared to medical experts [1]. However, current approaches classify not comprehensively for DR but only for specific DR symptoms. OCT Angiography (Figure 1) is an imaging technique, that enables a comprehensive DR diagnosis. However, due to its extensive image processing needs, datasets are only sparsely available and therefore seldomly used in ML work. In this work it is investigated, how raw, interferometric OCT data can be used for DR classification. In theory this could enable a comprehensive classification for DR while skipping image processing.
Methodology
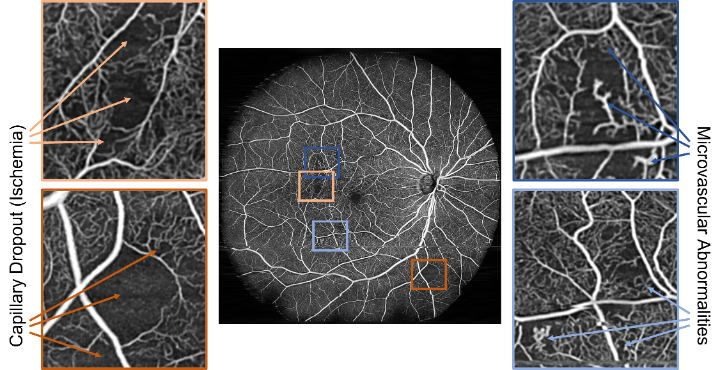
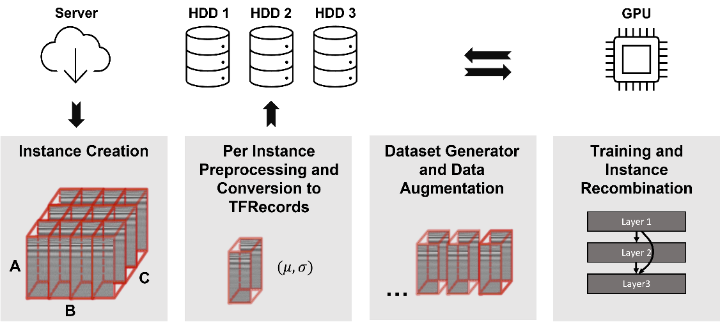
In order to achieve this, a Multiple Instance Learning (MIL) [2] approach is used. Since the raw, interferometric data is not humanly interpretable, the dataset cannot be manually annotated. Therefore, the diabetes status of patients was chosen as a weak label. Dataset samples (bags) were then split like a checkerboard into sub-patches (instances) which in turn inherited the annotation from their parent bags. This results in an inexactly labelled dataset of instances that satisfy the standard multiple instance (SMI) assumption. The full training workflow is shown in Figure 2. While training, the model’s performance is assessed only after recombining instances into their originating sample. This gets rid of the label noise resulting from inheriting labels to the instance level. For loss calculation, however, only the instance level was used since label noise does not negatively impact neural network performance. Lastly, the network’s performance was assessed by comparing Grad-CAM importance maps [3] with angiography images that were manually annotated by medical experts.
Image Results
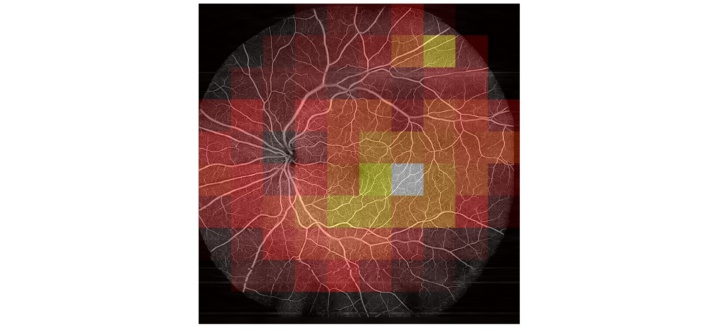
To verify that weak labels are sufficient for classifying the herein used dataset, in a first step OCT Angiography images were calculated from raw data and a classification network was trained on them. Using the MIL approach, a test accuracy of 93.3 % was reached. Early stopping with information from the bag level and data augmentation can significantly increase the model’s performance. Using the bag level for loss calculation significantly reduces the model’s performance. Comparing MIL to Single Instance Learning (SIL), only the MIL approach yields Grad-CAM features that correlate with actual DR pathologies (Figure 3). Using a transfer learning based approach yields grad-CAM features that seem similar to the MIL approach, albeit not as close to the manual annotations.
Raw Results
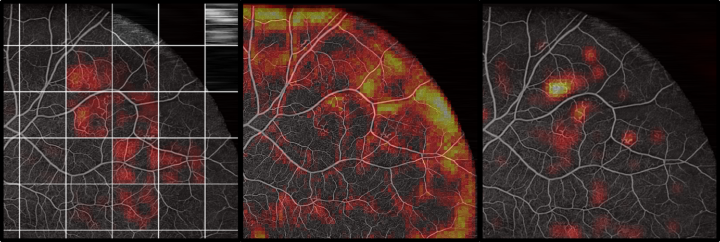
The raw dataset did only reach a low test accuracy of 58 %. Furthermore, areas that were predicted as very likely diabetic had no correlation to pathologies visible in the Angiography (Figure 4). This implies that the network learned to focus on patterns in the raw dataset, that do not generalize to an unknown dataset (like the actual pathologies would). Varying hyperparameters had only an insignificant influence. Several biases that exist in the dataset between the healthy and the diabetic class were detected and removed, but no increase in classification performance could be observed.
Conclusion
Generally, MIL and weak labels based on only the diabetes status are suitable for classification for DR. On the OCT Angiography dataset, results were better than using MIL than SIL, leading to the conclusion that MIL has a regularizing effect on training. On the raw dataset the network did either overfit on biases in the dataset, or on features that do not generalize. This leads to the conclusion, that the herein used dataset is not suitable for raw data classification.
References
[1] Feng Li et al. “Deep learning-based automated detection of retinal diseases using optical coherence tomography images”. In: Biomedical Optics Express 10 (2019). doi: https://doi.org/10.1364/BOE.10.006204.
[2] Jaume Amores. “Multiple instance classification: Review, taxonomy and comparative study”. In: Artificial Intelligence 201 (2013). doi: https://doi.org/10.1016/j.artint.2013.06.003.
[3] Ramprasaath R. Selvaraju et al. “Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization”. In: International Journal of Computer Vision (2016). doi: https://doi.org/10.1007/s11263-019-01228-7
Ansprechpartner

Uwe Sörgel
Prof. Dr.-Ing.Institutsleiter, Fachstudienberater